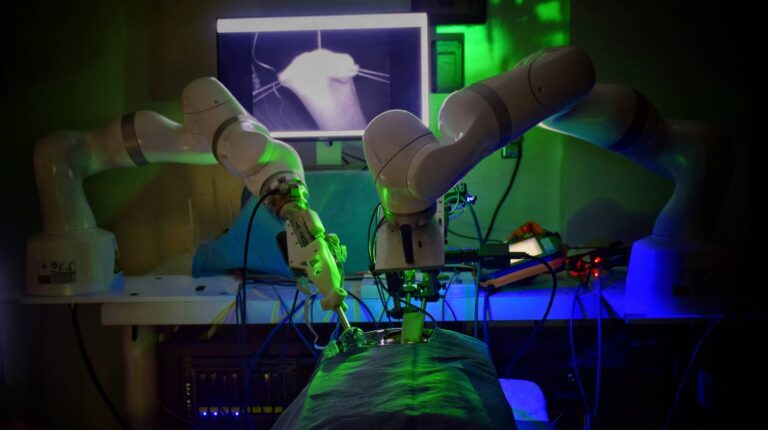
 Using machine learning and computer vision, a surgical robot successfully performs an anastomosis, demonstrating a notable step toward automated surgery.
Using machine learning and computer vision, a surgical robot successfully performs an anastomosis, demonstrating a notable step toward automated surgery.
In a medical first, a robot has performed laparoscopic surgery without the guidance of a surgeon’s hand. The study, recently publish in Science Robotics, outlines the design of an enhanced version of the Smart Tissue Autonomous Robot (STAR) that completed the challenging surgery on the soft tissue of a pig. The accomplishment marks a milestone toward fully automated robotic surgeries.
“Our findings show that we can automate one of the most intricate and delicate tasks in surgery: the reconnection of two ends of an intestine. The STAR performed the procedure in four animals and it produced significantly better results than humans performing the same procedure,” Axel Krieger, senior author and assistant professor of mechanical engineering at Johns Hopkins’ Whiting School of Engineering, said in a press release.
In laparoscopic procedures, surgeons use small incisions and a camera to perform an operation in the abdomen or pelvis. Anastomosis—which involves connecting two tubular structures such as blood vessels or intestines—is often performed laparoscopically. Despite being minimally invasive, the surgery has potential for serious complications to the patient if any leakage occurs due to flawed suturing.
Autonomous robotic surgery has the potential to improve medical efficiency, safety, and reliability. However, according to the study autonomous anastomosis poses challenges when it comes to intricate imaging, tissue tracking, and surgical planning. These procedures also often require quick adaptation if an issue arises during surgery.
The current STAR model improves on a 2016 iteration capable of suturing a pig’s intestine, however it required human intervention and created a larger incision.
With advanced robotic precision and suturing tools, along with a 3D imaging system and machine learning-based tracking algorithms, the latest STAR can adjust its surgical plan in real time.
“We developed machine learning, computer vision, and advanced control techniques to track the target tissue movement in response to patient breathing, detect the tissue deformations between different suturing steps, and operate the robot under motion constraints,” the researchers writer in the study.
A machine-learning algorithm based on convolutional neural networks (CNNs) predicts tissue motion and guides suture plans. The researchers trained the CNNs using 9,294 examples of motion profiles from anastomosis procedures, to learn tissue motion based on breathing patterns and other tissue motion during surgery.
The robot synchronizes with a camera to scan and create suture plans while the tissue is stationary. Using enhanced computer vision and a CNN-based landmark detection algorithm, STAR generates two initial suture plans to connect adjacent tissue. Once an operator selects a plan, the robot applies a suture to the tissue and reimages the area for tissue deformation.
If a change in tissue position is greater than 3 mm compared with the surgical plan, it notifies the operator to initiate a new suture planning and approval step. This process repeats for every suture.
According to Krieger, an NVIDIA GeForce GTX GPU was used for training and running the CNNs, including four convolutional, three dense layers, and two outputs that tracked tissue motion. Training and testing of the landmark detection algorithm, using a cascaded U-Net architecture, was performed with an NVIDIA T4 GPU.
The researchers examined the quality of the anastomosis, which includes needle placement corrections, suture spacing, size of suture bites, completion time, lumen patency, and leak pressure. They found the autonomous STAR outperformed the consistency and accuracy of both expert surgeons and robot-assisted surgeries.
“What makes the STAR special is that it is the first robotic system to plan, adapt, and execute a surgical plan in soft tissue with minimal human intervention,” Krieger said.
Read the study in Science Robotics. >>
Read more. >>



 Using machine learning and computer vision, a surgical robot successfully performs an anastomosis, demonstrating a notable step toward automated surgery.
Using machine learning and computer vision, a surgical robot successfully performs an anastomosis, demonstrating a notable step toward automated surgery. NVIDIA Jetson Nano is paving the way to detect certain types of cancer sooner.
NVIDIA Jetson Nano is paving the way to detect certain types of cancer sooner.